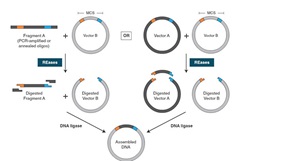
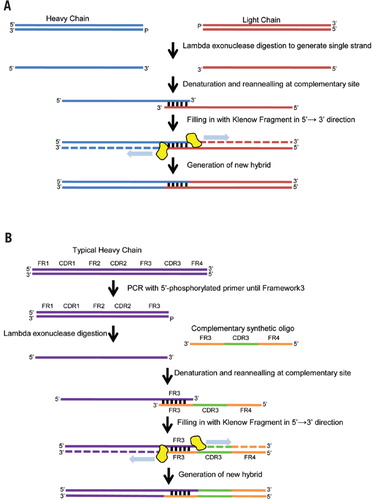
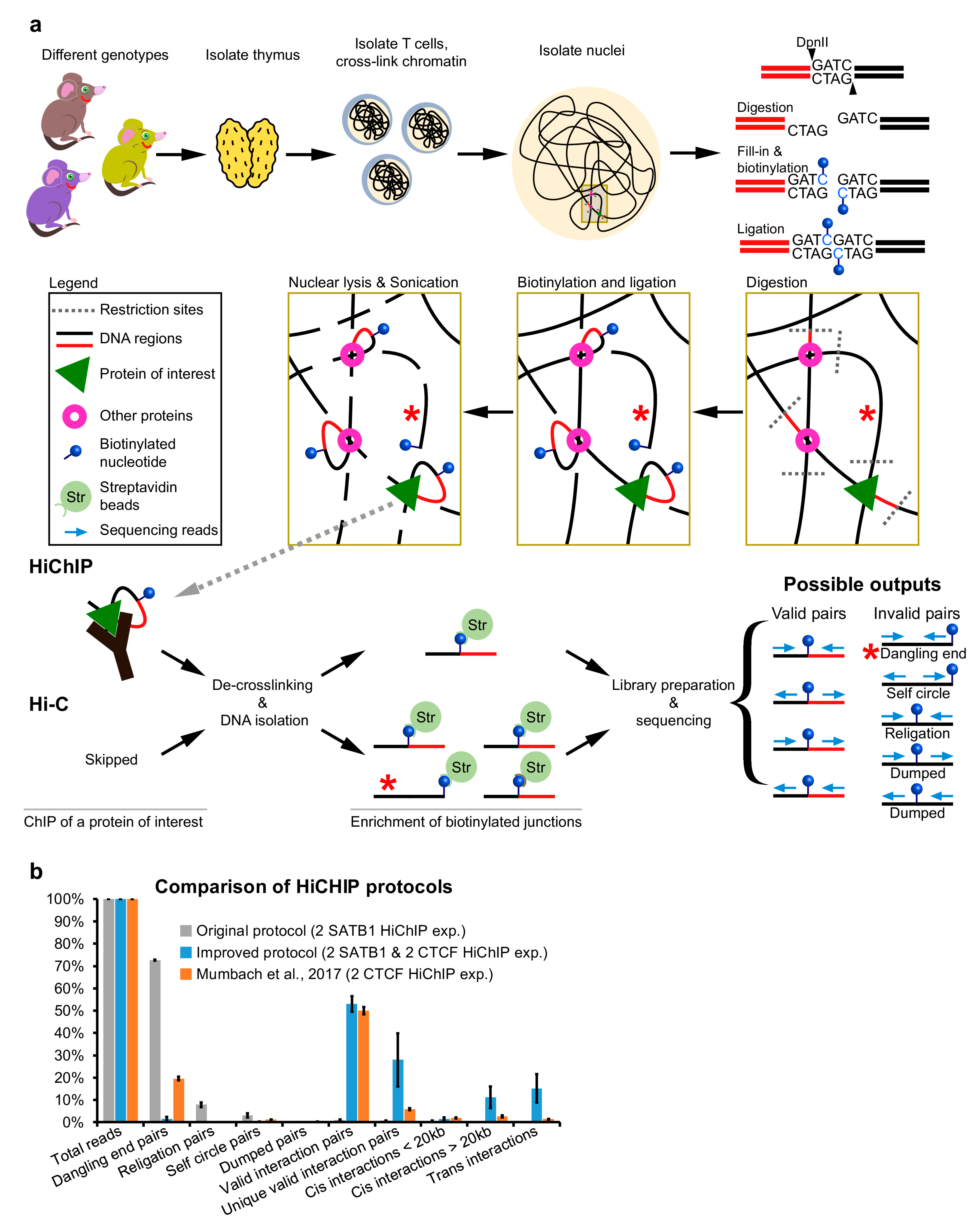
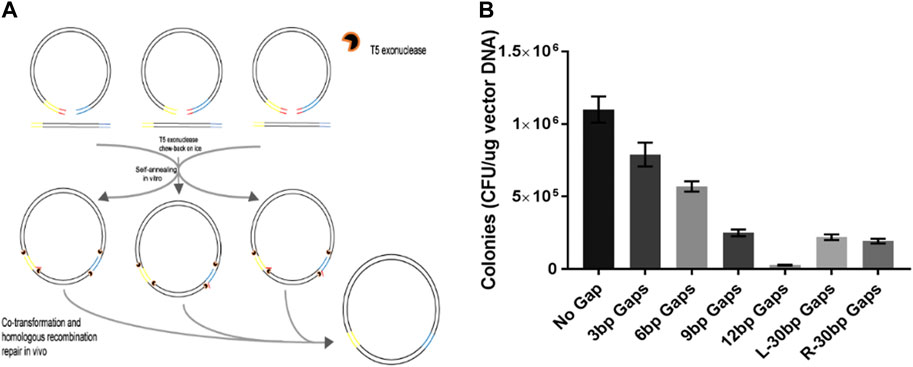
Schematic representation of the proposed protocol for generating new... | Download Scientific Diagram

Escherichia coli β-clamp slows down DNA polymerase I dependent nick translation while accelerating ligation | bioRxiv

Escherichia coli β-clamp slows down DNA polymerase I dependent nick translation while accelerating ligation | bioRxiv

DNA synthesis by Klenow fragment exonuclease minus (KF exo-) on a DNA... | Download Scientific Diagram
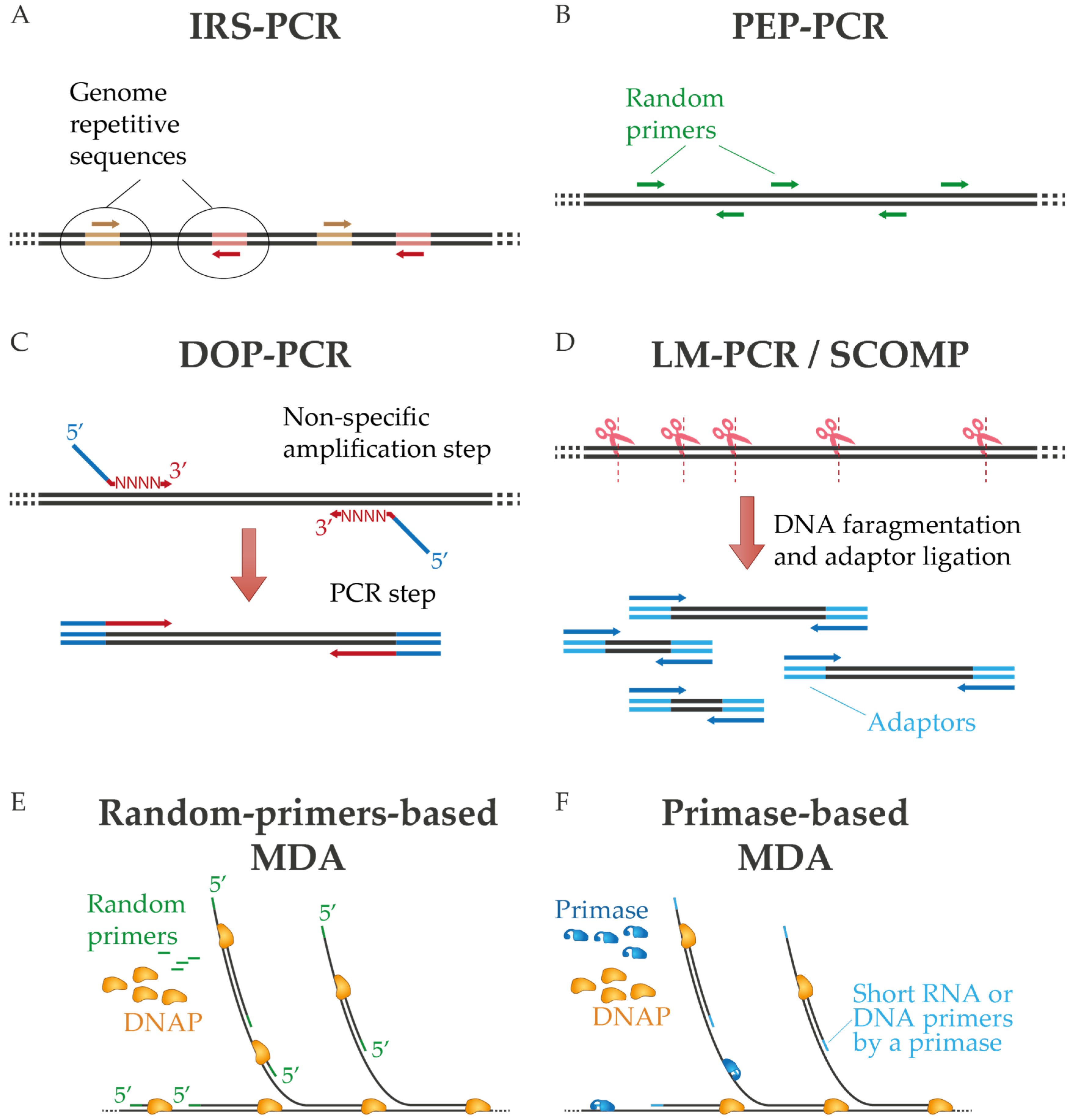
IJMS | Free Full-Text | DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions

Structure of the Herpes Simplex Virus 1 Genome: Manipulation of Nicks and Gaps Can Abrogate Infectivity and Alter the Cellular DNA Damage Response | Journal of Virology
Escherichia coli β-clamp slows down DNA polymerase I dependent nick translation while accelerating ligation | PLOS ONE
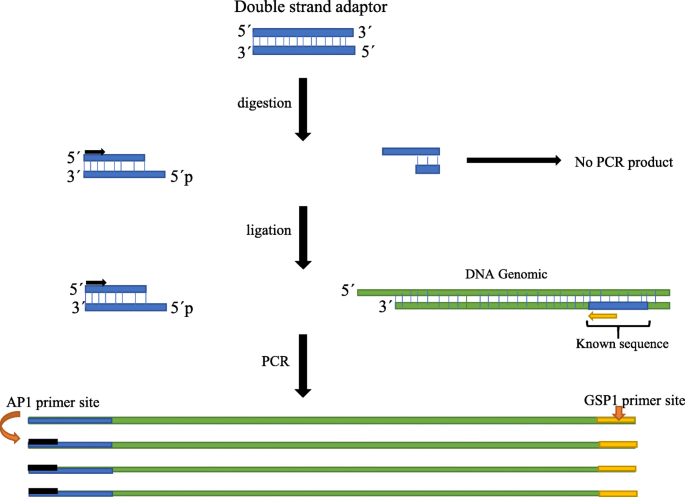
Simple innovative adaptor to improve genome walking with convenient PCR | Journal of Genetic Engineering and Biotechnology | Full Text

DNA Ligase C and Prim-PolC participate in base excision repair in mycobacteria | Nature Communications

DNA Ligase C and Prim-PolC participate in base excision repair in mycobacteria | Nature Communications

A multifunctional DNA polymerase I involves in the maturation of Okazaki fragments during the lagging‐strand DNA synthesis in Helicobacter pylori - Cheng - 2021 - The FEBS Journal - Wiley Online Library

WO1993000447A1 - Amplification of target nucleic acids using gap filling ligase chain reaction - Google Patents

Structural Transformation of Wireframe DNA Origami via DNA Polymerase Assisted Gap-Filling | ACS Nano
Outline of in vivo and in vitro gap-filling assays. (A) The in vivo... | Download Scientific Diagram